Highly potent and selective PPAR delta agonist reverses memory deficits in mouse models of Alzheimer's disease.
Kim, H.J., Kim, H., Song, J., Hong, J.Y., Lee, E.H., Londhe, A.M., Choi, J.W., Park, S.J., Oh, E., Yoon, H., Hwang, H., Hahn, D., Jung, K., Kwon, S., Kadayat, T.M., Ma, M.J., Joo, J., Kim, J., Bae, J.H., Hwang, H., Pae, A.N., Cho, S.J., Park, J.H., Chin, J., Kang, H., Park, K.D.(2024) Theranostics 14: 6088-6108
- PubMed: 39431021
- DOI: https://doi.org/10.7150/thno.96707
- Primary Citation of Related Structures:
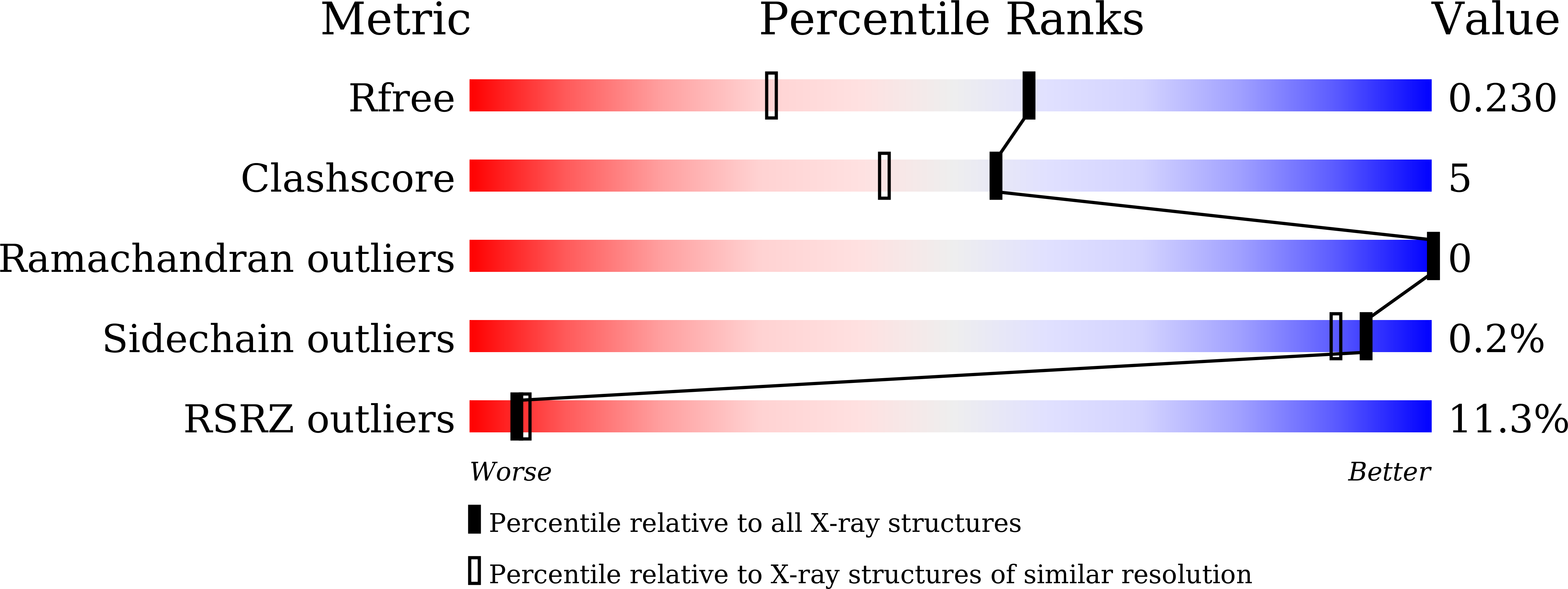
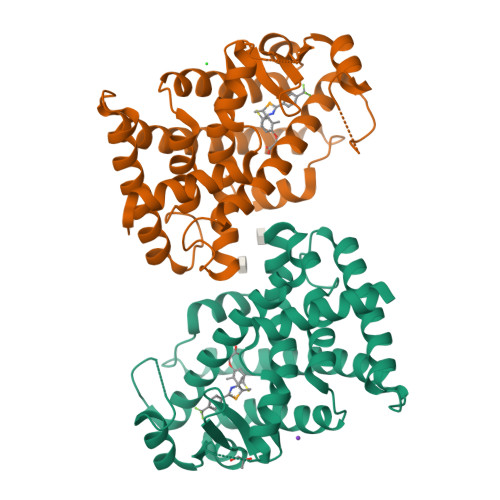
5Y7X - PubMed Abstract:
Rationale: Alzheimer's disease (AD) is a progressive neurodegenerative disease accompanied by neurotoxicity, excessive inflammation, and cognitive impairment. The peroxisome proliferator-activated receptor (PPAR) δ is a potential target for AD. However, its regulatory mechanisms and therapeutic potential in AD remain unclear. We aimed to investigate if the activation of PPARδ using a highly selective and potent agonist could provide an effective therapeutic strategy against AD. Methods: We synthesized a novel PPARδ agonist, 5a, containing a selenazole group and determined the X-ray crystal structure of its complex with PPARδ. The drug-like properties of 5a were assessed by analyzing cytochrome P450 (CYP) inhibition, microsomal stability, pharmacokinetics, and mutagenicity. We investigated the anti-inflammatory effects of 5a using lipopolysaccharide (LPS)-stimulated BV-2 microglia and neuroinflammatory mouse model. The therapeutic efficacy of 5a was evaluated in AD mice with scopolamine-induced memory impairment and APP/PS1 by analyzing cognitive function, glial reactivity, and amyloid pathology. Results: Compound 5a , the most potent and selective PPARδ agonist, was confirmed to bind hPPARδ in a complex by X-ray crystallographic analysis. PPARδ activation using 5a showed potent anti-inflammatory effects in activated glial cells and mouse model of neuroinflammation. Administration of 5a inhibited amyloid plaque deposition by suppressing the expression of neuronal beta-site amyloid precursor protein cleaving enzyme 1 (BACE1), and reduced abnormal glial hyperactivation and inflammatory responses, resulting in improved learning and memory in the APP/PS1 mouse model of AD. Conclusion: We identified that specific activation of PPARδ provides therapeutic effects on multiple pathogenic phenotypes of AD, including neuroinflammation and amyloid deposition. Our findings suggest the potential of PPARδ as a promising drug target for treating AD.
Organizational Affiliation:
Center for Brain Disorders, Korea Institute of Science and Technology (KIST), Seoul 02792, Republic of Korea.